
Unraveling the genes responsible for late flowering in the giant woody cabbage (Brassica oleracea).
Backgroud& context
Why are there so many phenotypic differences between woody and herbaceous species? To this day, scientists have failed to provide satisfying answers. We have crossed a tall, woody, late flowering cabbage accession (Jersey kale) with a small, rapid flowering cabbage accession (TO1000) and obtained 200 F2 accessions in tissue culture (see picture), allowing multiple experiments of the same genotype over time. This project focuses on the F1 generation that displays a wide difference of flowering time, ranging from 4 to 17 months after potting the in-tissue culture. This variation in F1 is due to the heterozygosity of the Jersey kale parent. In the meanwhile, we have generated a next-generation sequencing dataset and performed preliminary analyses to uncover the genes involved in flowering time.
More information is found on the website of the University of Leiden


Fig. 1. Left: in-tissue culture of three accessions: TO1000, one of the F1 accessions, and the Jersey kale parent. Right: the same three accessions in mature state.
Objectives and goals
Based on an existing next-generation dataset, this MSc project wants to unravel the genes responsible for flowering time in Brassica oleracea.
Material, tasksand approach
This MSc proposal wants to build on a previous Bulk Segregant Analysis (BSA) where we observed genomic differences between four F1 populations using the R package created by [1]. Therefore, we pooled DNA in four groups based on flowering time, and performed next-generation sequencing. The four groups are: early flowering (flowering 113-135 days after potting), intermediate flowering (flowering 154-163 days after potting), late flowering (flowering 176-239 days after potting), and the genotypes that did not flower at the onset of DNA extraction (all but one of these genotypes did flower later, up to 533 days after potting).
The preliminary BSA analyses revealed interesting genomic variation between the pooled groups, especially in chromosome 6 (Fig. 2). This variation will be confronted with the pathway variation for flowering time genes in the Brassica rapa genome based on a published study ([2], Fig. 3).
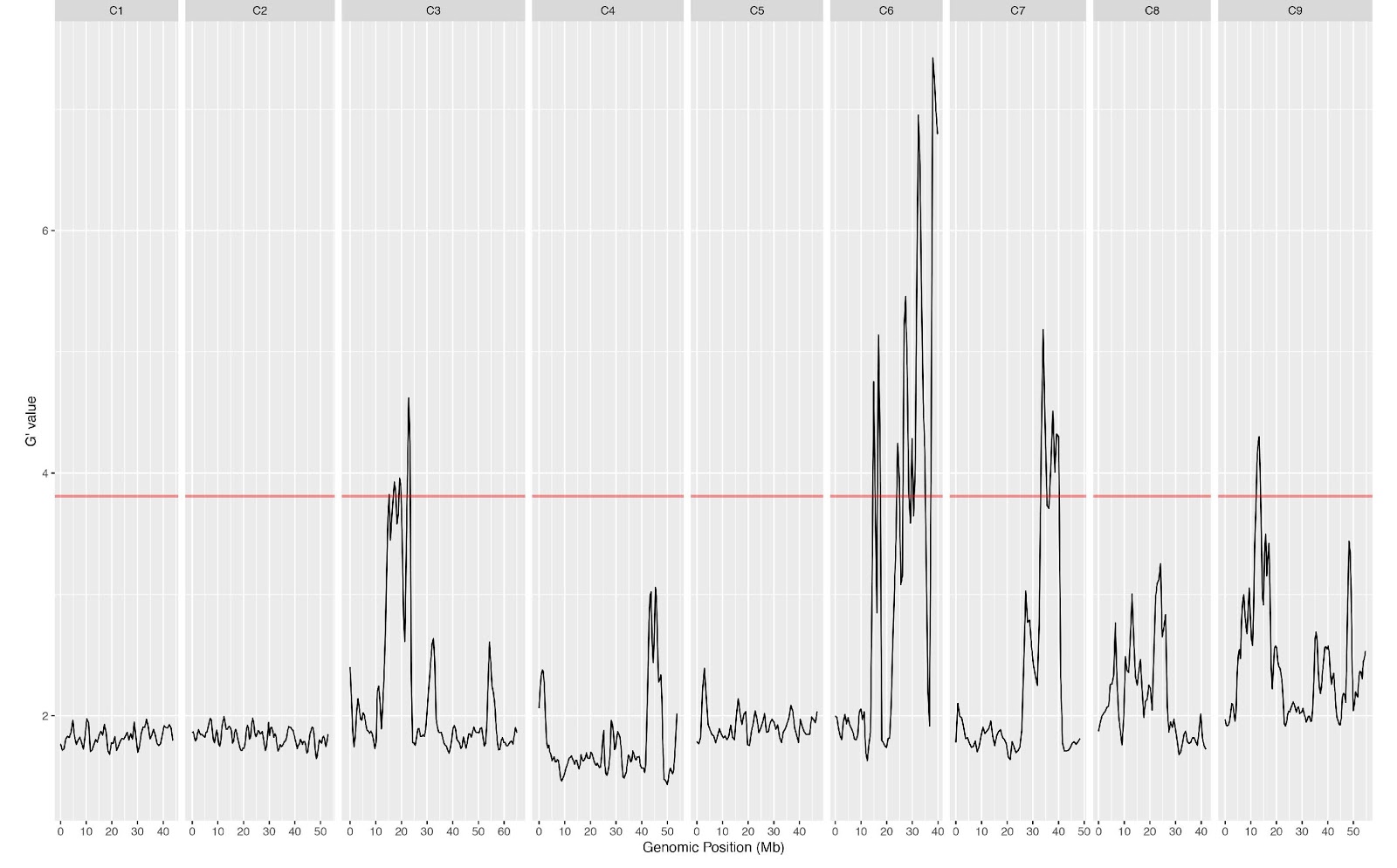
Fig. 2: Outcome of one of the BSA analyses based on one group comparison, showing interesting potential candidates involved in wood formation on chromosome 6.
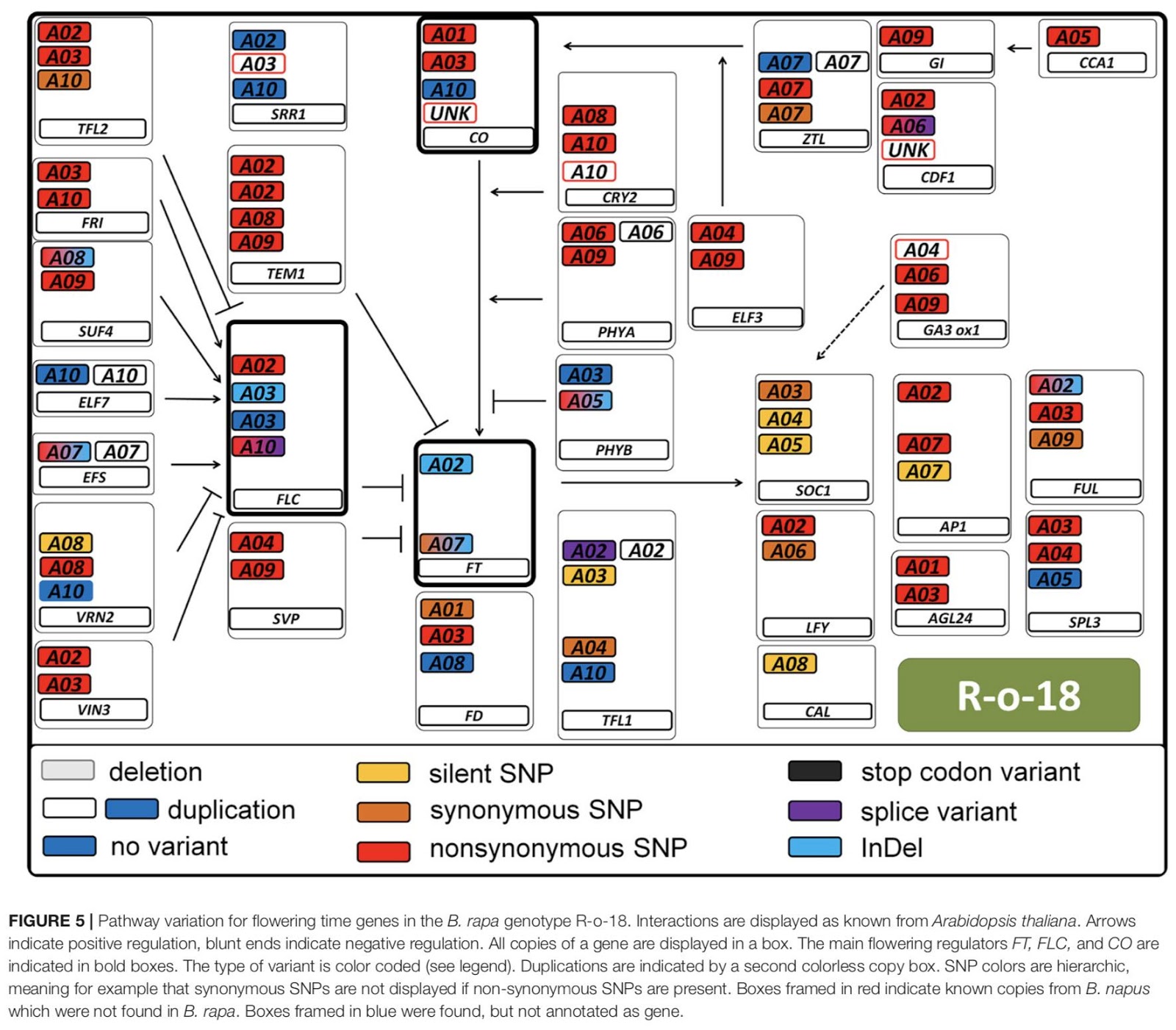
Fig. 3. Pathway variation for flowering time genes in the Brassica rapa genome. Arrow point to positive regulation, blunt ends indicate negative regulation. All copies of a gene are displayed in a box. The main flowering regulators FT, FLC and CO are indicated in bold boxes (from [2]).
Studentrequirements
A motivated MSc student in bio-informatics who is interested in the genetic mechanism behind flowering time and has sufficient writing skills to contribute to a scientific publication.
Projectreferences
[1] Mansfeld and Grumet. 2018. QTLseqr: an R package for Bulk Segregant Analysis with next-generation sequencing. Plant Genome 11:180006.
[2] Schiessl SV, Huettel B, Kuehn D, Reinhardt R, Snowdon RJ. 2017. Flowering time gene variation in Brassica species shows evolutionary principles. Frontiers in Plant Sciences 8: 1742.